A great presentation by our Jeev on the use of Flyte at Freenome. He presented in the Flyte OSS Community meeting and it was recorded. Check it out
Job Opportunities for Recent Graduates at our Freenome Research Platform
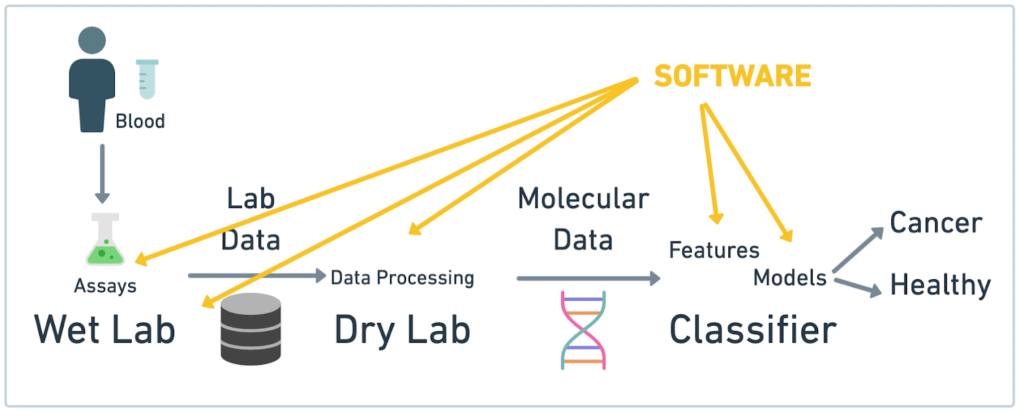
Freenome combines molecular biology and machine learning to create tools that empower everyone to prevent, detect, and treat their disease. We are growing fast and we are hiring in many areas; and I have several openings for Recent Grads in my group.
Our group builds the lower layers for the Research Platform used by all our scientists. Our focus is on the compute, data, automation and reliability aspects of the platform. We use Python, multiple GCP services, k8s, workflows (go flyte!) and other modern frameworks and tools. We collaborate daily with engineers that are building bioengineering and machine learning tools used to ingest and process data and run multiple types of experiments
You do not need to know molecular biology – I knew nothing when I arrived – but you will learn a lot. You will also learn how a modern MLOps is put together. This is a very special opportunity to make a difference and to develop your technical skills.
We have a great technology, our Multiomics platform, a great multi-cultural team, last time I counted over 31 languages at Freenome, great collaborators, partners and investors, see our News Page, and our mission is truly unbeatable: let’s solve cancer together!
We are looking for somebody with a BS or higher in Computer Science, Data Engineering, or related field or equivalent training, fellowship and/or work experience. The majority of our work is based on Python so that is strongly recommended. You will work in an agile development cycle, interacting with multiple groups; your written and verbal skills will help you accomplish your tasks. We emphasize collaboration, communication and compromise and leverage our collective skills to achieve our goal.
If you are interested in learning about this opportunity, start by checking if you know somebody at Freenome that graduated at your school and talk with them. If you think there may be a fit, you can ask them to refer you at the internal job site. Or ping me if interested (LinkedIn or mail); we know graduations are now and good candidates will have multiple opportunities, so we want to move quickly.
And yes, we are open to remote work, though only in the US at the moment.
You, your Career and BioTech
Sometimes it is easy to forget that your job and your career are two different things. Your job is what you do for your employer. Your career is the path that will get your through a sequence of jobs to achieve your personal goals. Your career will be influenced by events outside of your control, like life events or the economy, but you do have some control over your career and you should use it.
A few weeks ago I gave a presentation on this topic to the CS Seniors at CSUM – Cal State University Monterrey Bay – as part of a series on Life After College. The short summary of the presentation was:
- Your Job is not your Career – You are in charge of your Career,
- Software is a key part of BioTech,
- You should consider adding BioTech to your career Choices.
During the presentation I used as an example Freenome, an early stage BioTech company that I joined at the end of 2020 after spending most of my career in HiTech.
I re-recorded the presentation into a 9 minute video that you can see HERE.
Enjoy and stay connected!
And Freenome is Hiring!
Change is Good – 11 years after Sun Microsystems
Change is Good!
The acquisition of Sun Microsystems by Oracle Corp closed 11 years ago, on Jan 27th, 2010, 9 months after it was first announced.
I had been at Sun for many years and I really liked what I was doing but, in hindsight, the acquisition forced me to accept change, and that was good. I stayed at Oracle for a few months, and then left for BlackBerry (then RIM) where I joined other ex-Sun people, learned new things and shared some of what I had learned with new colleagues in a different community.
I believe that embracing change, and exchanging communities, is a key asset in our industry. We learn new things through experimentation and observation, and then reshuffle and try again in a new environment. And, collectively, we move forward.
I reported on the beginning of the Sun exodus a year after the acquisition: people and OSS projects. The process has continued, though I have only tracked the movement through my direct friends. On a personal level, after BlackBerry I joined Progress, and then CloudBees.
I’m now at Freenome. I joined for three main reasons:
- Cancer sucks and I believe I can help Freenome in its mission.
- I can learn new things: Biology, ML, Workflows, Python, Continuous Improvement in a Regulated Industry, and many other topics
- The company culture is outstanding
The change to Freenome is arguably the biggest change so far for me, from hiTech to bioTech. This change enables many opportunities for learning and teaching across these two industries, and I will write about that in the future but, in the meantime…
Change may sometimes be scary but, change is good! My lesson is to embrace it and keep growing…
And ping me if you are considering a move to BioTech. Particularly if Freenome has any appeal to you.

Freenome Bloggers
A list of Freenome people writing about Freenome events and content.
Freenome/Blog
A collection of links to blog posts from Freenome people about Freenome…
- Picnic 2018 – Working at Freenome: Sometimes it’s a picnic!
- Back to the future with the Freenome Holiday Party
- Freenome’s 3rd Annual Picnic: Team building with bubbles!
- Uniting in Monterey for some fun in the sun! — Freenome
- Freenome and The Women’s Building Volunteer Event
- Service, it’s in our blood — Freenome
- Freenome event supporting women in STEM fields
Teddy
- (Dec ’18) Freenome: Where I’ll Be This Summer!
- (Nov ’19) Freenome: Summer Experience and Where I’ll be Next Year
- (Feb ’21) SWE at Freenome. My (Almost) First Year at Freenome
Eduardo
- (Feb ’21) Juggling at Freenome, and…
- Other Posts
CRC and Freenome
A collection of links to and extracts from public information on CRC and Freenome (last updated on Feb 14, 2021). Also see similar post collections on Social Topics, and Technology/Science.
Disclaimer: I am a Software guy, barely beginning to learn the Biology side of all this :). Plus this post is WiP
From Wikipedia: “Colorectal cancer (CRC) is the development of cancer from the colon or rectum“. Also see American Cancer Society and Mayo Clinic. Wikipedia maintains a List of (Public) People Diagnosed with CRC; a notable recent death was that of Chadwick Boseman.
Unless indicated otherwise, the links below are to Freenome Press Releases with an excerpt and an occasional brief commentary.
The current lack of an effective, blood-based test may be a contributing factor to low rates of CRC screening today, with nearly 1 in 3 adults aged 50-75 years non-adherent with screening guidelines. Existing methods, such as colonoscopy and stool-based testing, are often seen as inconvenient and uncomfortable by patients, leading many to postpone or forgo recommended screenings
A concise description of the value of a blood-based screening for CRC.
and
We have shown that it’s possible to take a machine learning–based approach to decode the relationship between a patient’s cell-free DNA profile and his or her cancer status, detecting CRC in our dataset with a top performance of 82% sensitivity at 85% specificity. These are very encouraging data that support the continued development of a CRC screening test that includes cfDNA and machine learning as key components.
Above is a quote from Freenome’s Chief Medical Officer, Girish Putcha. Sensitivity is a measure of how well the test can identify true positives, while Specifity is a measure of how well the test can identify true negatives (see Wikipedia). There is a trade-off between sensitivity and specificity and, as indicated in public reports elsewhere one of the key benefits of the Freenome approach is that we can tune the tradeoff depending on the cancer and the treatments.
Freenome’s multiomics blood test can detect early-stage colorectal cancer (stage I/II) at 94% sensitivity and 94% specificity.
The improved sensitivity and specificity above numbers reflect advances since the earlier results reported in Oct 2018. The test reported here combines cell-free DNA (cfDNA) as well as protein biomarkers into a machine-learning classifier.
Following strong data presented at ASCO-GI … we are initiating the pivotal trial for our blood-based CRC test
Initiation of the CRC Clinical Trial
PREEMPT CRC is Freenome’s prospective clinical trial that will enroll 14,000 individuals to validate its multiomics blood test for CRC screening and FDA approval. The study will include asymptomatic, average-risk patients between the ages of 45 and 85 undergoing a screening colonoscopy.
And some details of the CRC Clinical Trial.
(Jan ‘21) Freenome’s Multiomics Blood Test Shows Promising Results in Detecting Colorectal Advanced Adenomas
multiomics blood test can detect colorectal advanced adenomas at 41% sensitivity and 90% specificity,
Concise description of the result.
The data from a pre-defined subset of AI-EMERGE® (n=522) showed that Freenome’s novel multiomics blood test for colorectal cancer screening was able to detect colorectal advanced adenomas (AAs) with a sensitivity of 41% at a specificity of 90%. Compared with the FDA-approved mSEPT9 (methylated septin 9) blood test, Freenome’s multiomics blood test showed much higher sensitivity (41% vs. 22%)1 for detecting AAs. When compared to currently available stool-based tests, the test demonstrated much higher AA sensitivity than a fecal immunochemical test, or FIT (41% vs. 24%) and comparable AA sensitivity to FIT-DNA (41% vs. 42%)2
And more details. Note that, unlike the report from 2020, this one is reporting on Advanced Adenomas, i.e. pre-cancer.
Freenome is Hiring!
Freenome is hiring!
The mission can’t be better…

The technology stack is mostly Python in the back-end, on K8s and GCP (et al.), and React in the front-end. I’ll provide more insight as I ramp up in 2021 but you can look at the openings in the Freenome career’s page. There are two relevant groups of openings, one for Computational Science and one for Engineering – I am in the Engineering group, and I’d expect most of my friends/contacts to be in that area but look around. The core approach is multiomics, using molecular biology and machine learning to detect cancers in blood, so regardless of where you land, Freenome would give you an opportunity to learn about those two areas.
The company’s HQ is in South San Francisco, in the Verily campus which includes the usual Alphabet goodies. We are all working from home right now, and there is some remote work flexibility.
… and now at Freenome!
Today (Dec 28th), I started at Freenome. Freenome is developing and productizing a Multiomics platform for early cancer detection. The tag line is “Spot the pattern, treat the cancer“. Multiomics means doing analysis on data from multiple “omes“, i.e. using fragments of DNA, RNA, proteins and other biomarkers from blood plasma. The first study is PREEMPT CRC – Colorectal Cancer.
I am joining as an Engineering Manager, same role as what I had at CloudBees. The technology platform is mostly Python in the back-end, plus JS in the front-end, plus… I’ll find out.
I’m very excited to join Freenome. I liked the team and the company, and loved the mission! I’ve lost multiple friends and relatives to Cancer, and I myself had an early encounter with Prostate Cancer, so any little thing I can do to help, sign me in!
Freenome is located in South San Francisco, close to many other BioTech companies, and has a strong list of partners and investors. In the short-term, I will be working from home – like everybody else.
Some of my reading material 🙂

CloudBees – Time to move on!
A delayed news update …
I left CloudBees last month. It was a great journey. We started with a basic concept, a very small team and somewhat limited experience with modern SaaS. We ended with two multi-featured products (DevOptics and SDM), on AWS and GCP, in a multi-region team, releasing in a CI/CD workflow using a very modern stack. We all learned a lot about technology and processes.
And great people! Smart, kind, team players, with many accents and cultures distributed all over. In the US we had people in the West (CA, OR, WA) and East Coasts (PA, MA, NC, FL) – and Texas, let’s not forget Texas! Outside the US, we had people in Australia (yep, 6 hours behind me) Ireland, Scotland, and England, Spain, France, Austria, Switzerland, (that’s CET for you, 9 hours ahead of me), and briefly Croatia (that’s 10 hours ahead!) – I became fairly good at Time-Zone math 🙂 And I learned about Irish and Australian sense of humor…
Thanks to everybody that shared this journey with me; do stay in touch.
CloudBees was great but it was time for me to move on.
I took a short break between jobs but I start at my new job tomorrow. More about it tomorrow!
Modeling Value Streams using DevOptics Free Plan
Hot off the press: you can now model your value stream and track your deployment frequency using the Free Plan for CloudBees DevOptics. We released this new functionality as part of the ramp-up to next week’s DevOps World | Jenkins World San Francisco. There are two main new aspects: an improved, and free, value stream modeler, and extending the availability of deployment frequency available to the Free plan.
With the stream modeler you can capture how software flows through your system (or you can design how you want it to flow) by creating and connecting Gates. Then, if your flow is based on jobs running on Jenkins or CloudBees Core CI/CD masters, you can connect these gates to the jobs using the DevOptics Plugin and leverage the computational aspects of CloudBees DevOptics.
The connected masters need to be configured to a DevOptics account, either in a Free plan or in a Premium plan. Both plans provide CD platform monitoring (Run Insights) – the Free plan only provides 7 days of statistics while Premium extends that to 90 days. The Free plan now also provides Deployment Frequency metric for all gates, albeit also only for 7 days. The Premium plan adds two more Job-based metrics: Change Failure Rate and Mean Time to Recovery as well as Mean Lead Time. Mean Lead Time is based on Value propagation in the form of commits and tickets through your Value Stream, which is a key feature of DevOptics Premium.
You can get more details about all this from the CloudBees DevOptics User Guide, and you can try it out from devoptics.cloudbees.com!
Using the Value Stream modeler is straight-forward. Setting up the plugin to enable Deployment Frequency is pretty easy too but below I spell out the steps, starting right after my earlier post: Upgrading to the latest DevOptics…
After connecting the plugin go to the Settings creen in DevOptics This is how it looks if you have configured only have Run Insights section of the Plugin:
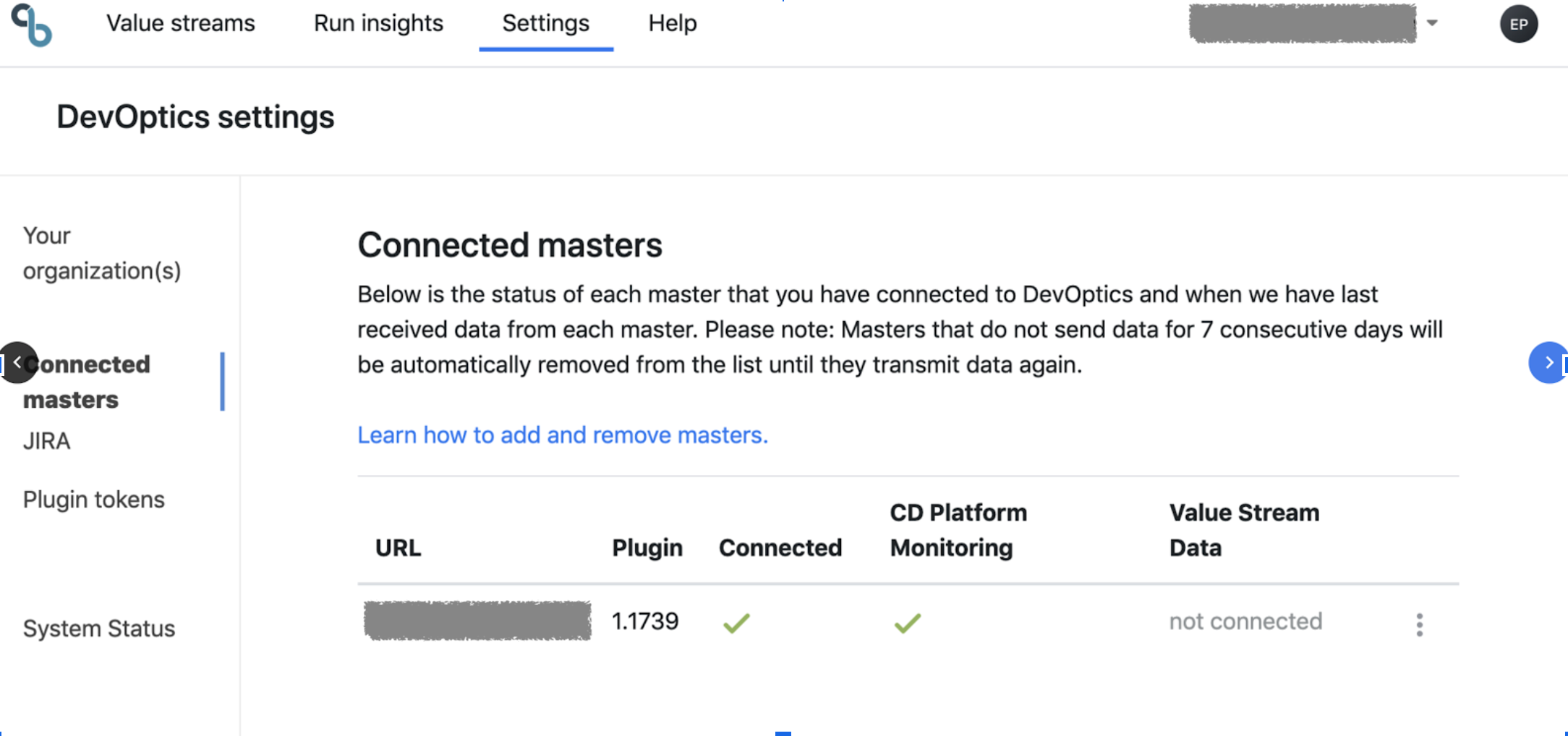
And this is how it looks after connecting also the Value Streams section:

Now create your value stream:
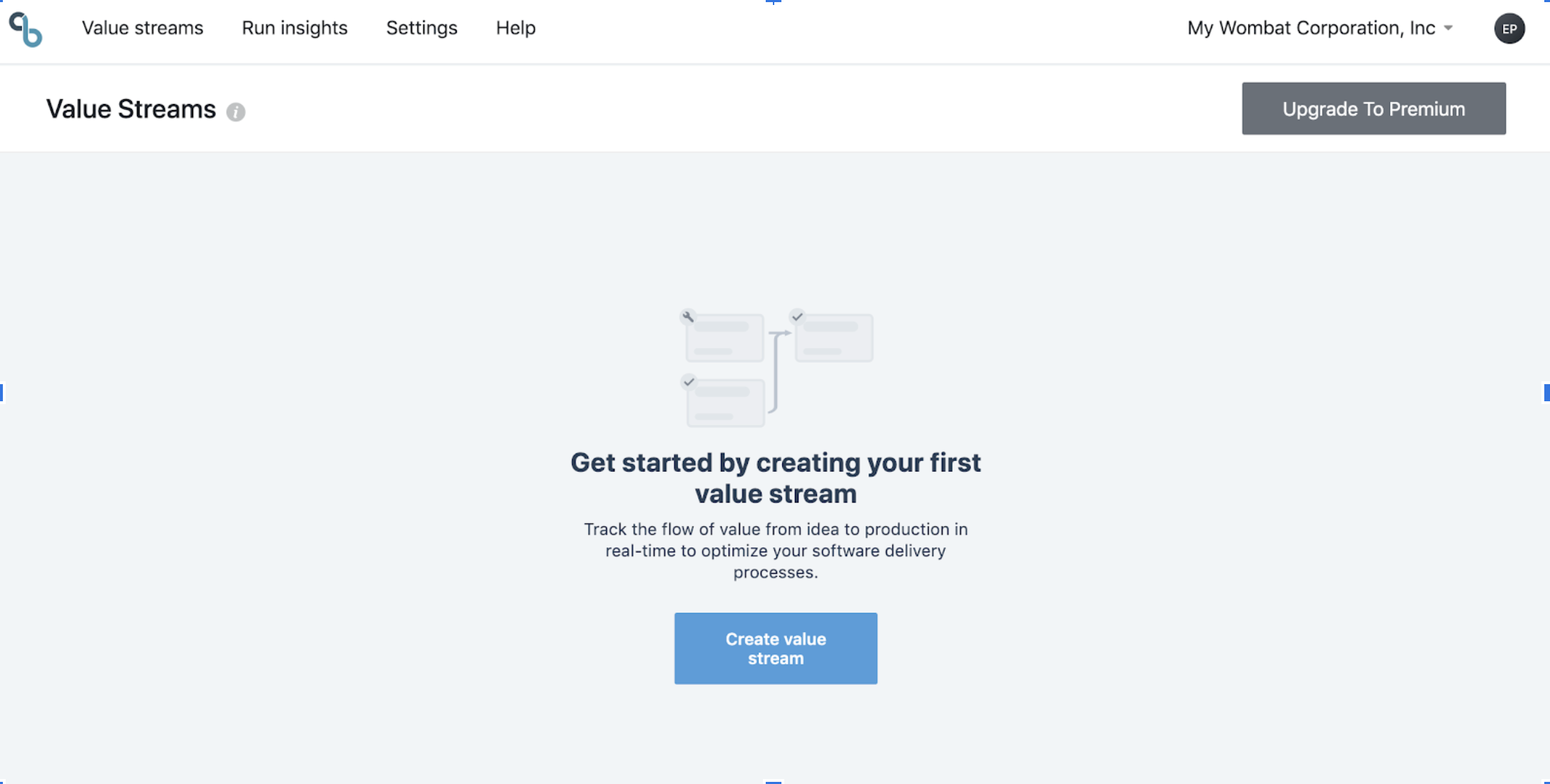
Here is a very simple one-gate value stream
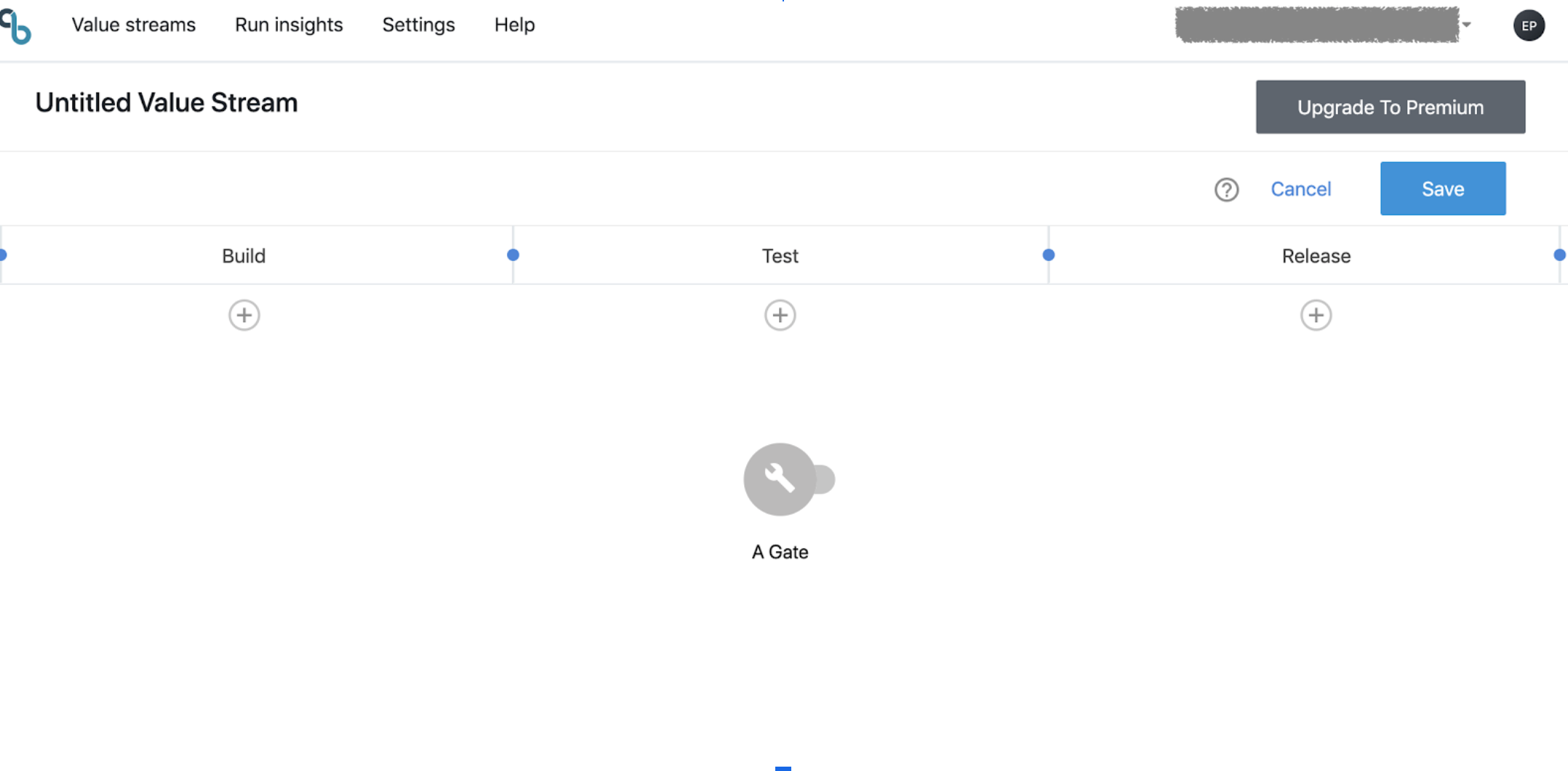
If we leave the gate unconfigured, it will look as this (note the wrench icon):
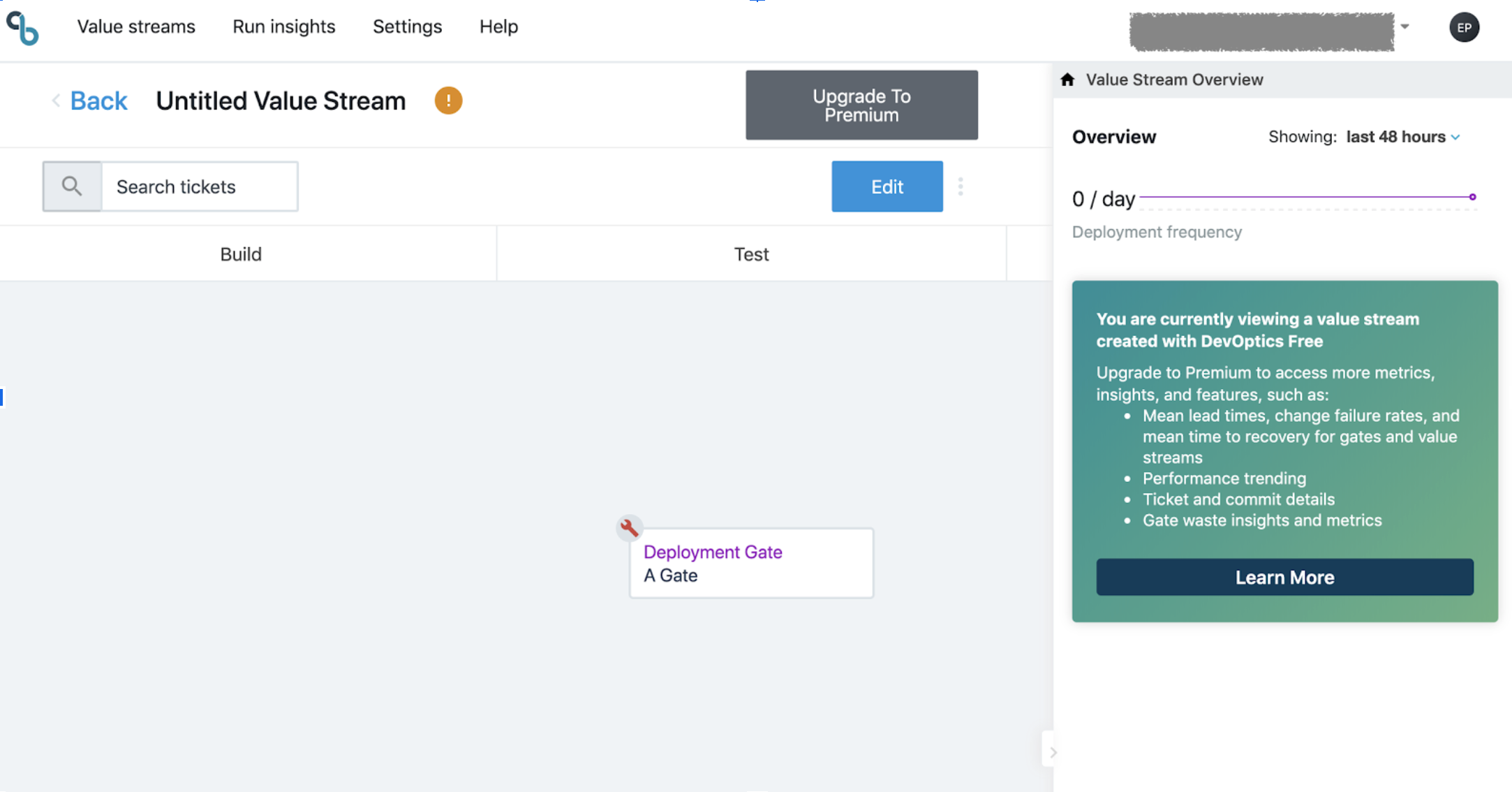
But, since we want to track Deployment Frequencies, we will now edit and configure the gate against one of the jobs in a master where a plugin has been installed. This is the configuration popup:

Save and you will see that the wrench goes away:
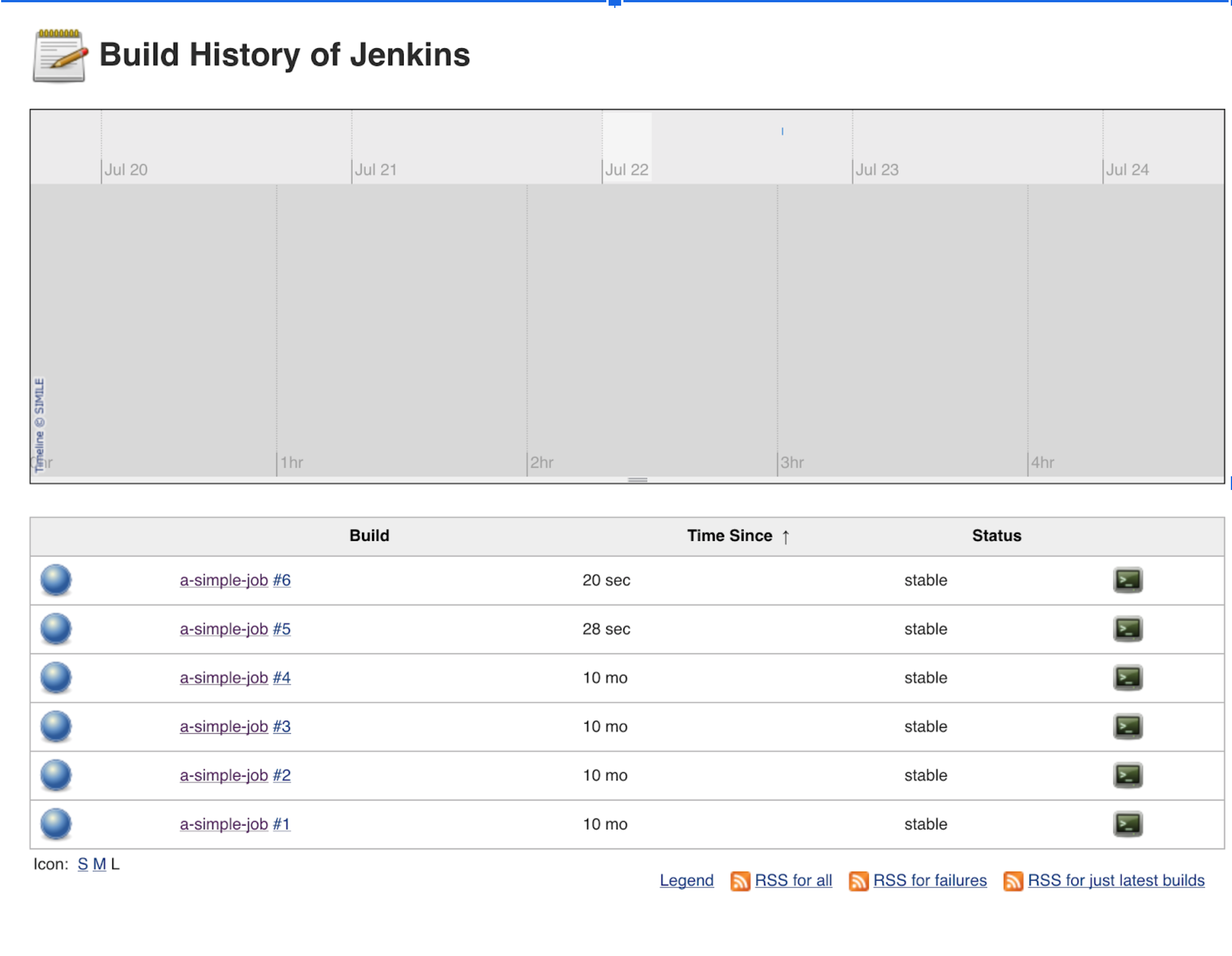
At this point we are ready! We just need to run some jobs on the master and data will flow, so…
We go to our Jenkins and run some jobs…
Now if we go back to the DevOptics app and look at the Value Streams screen we will see something like this:
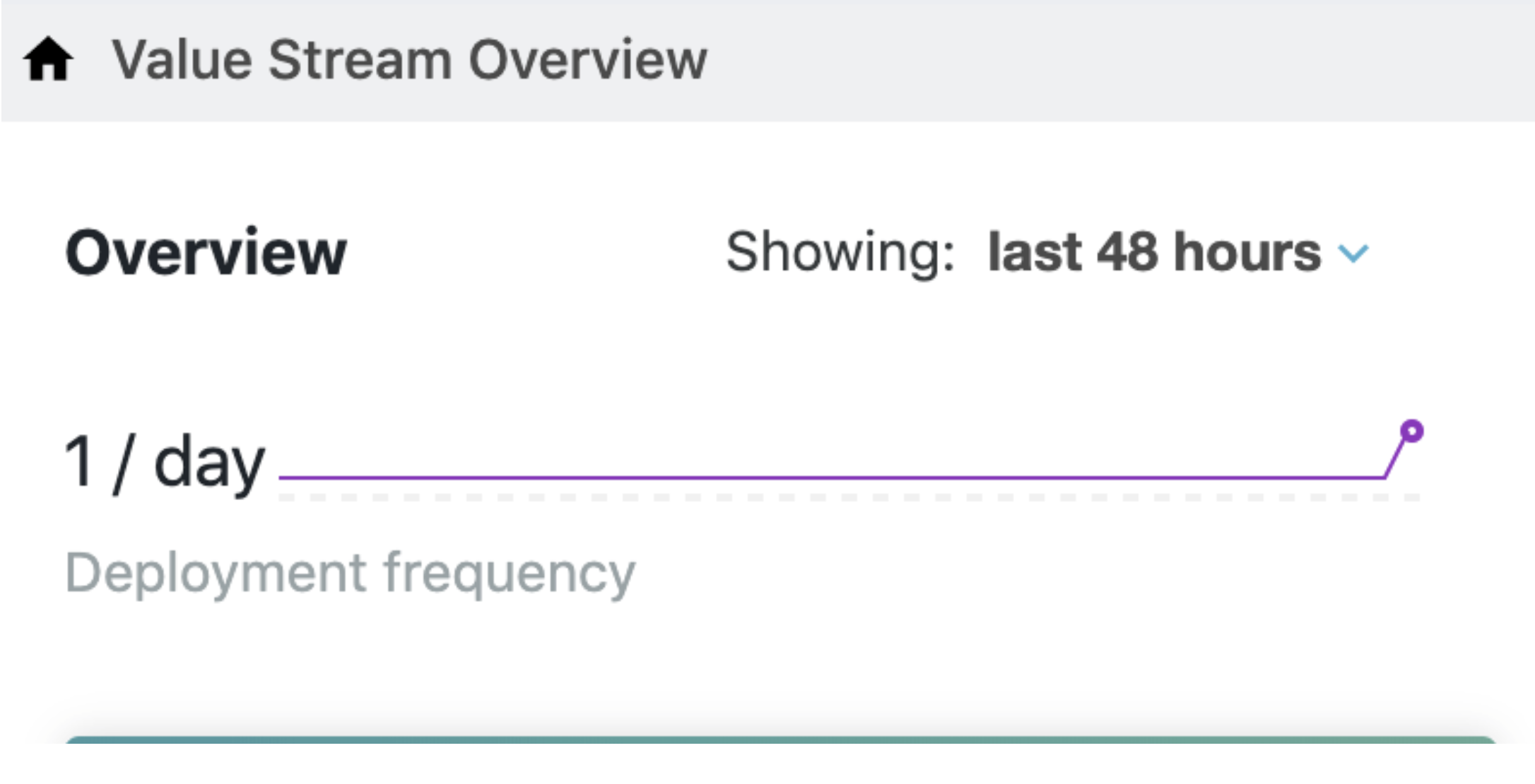
On the Run Insights side, the graphs will initially look as below – note that the grey bars are gone and there is some empty space for the most immediate time interval:
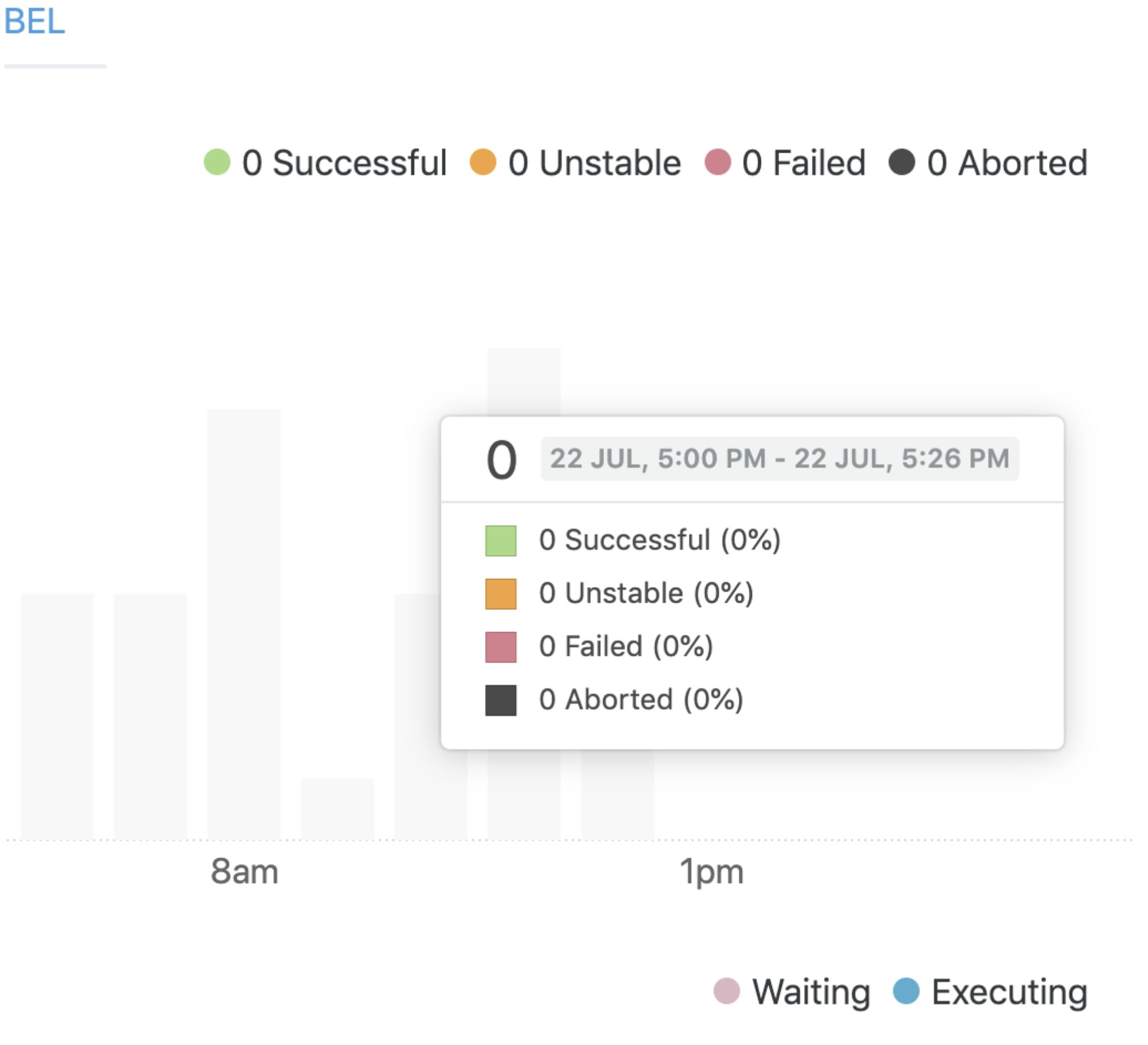
But, if we wait a bit for the data to be propagated, we will see:
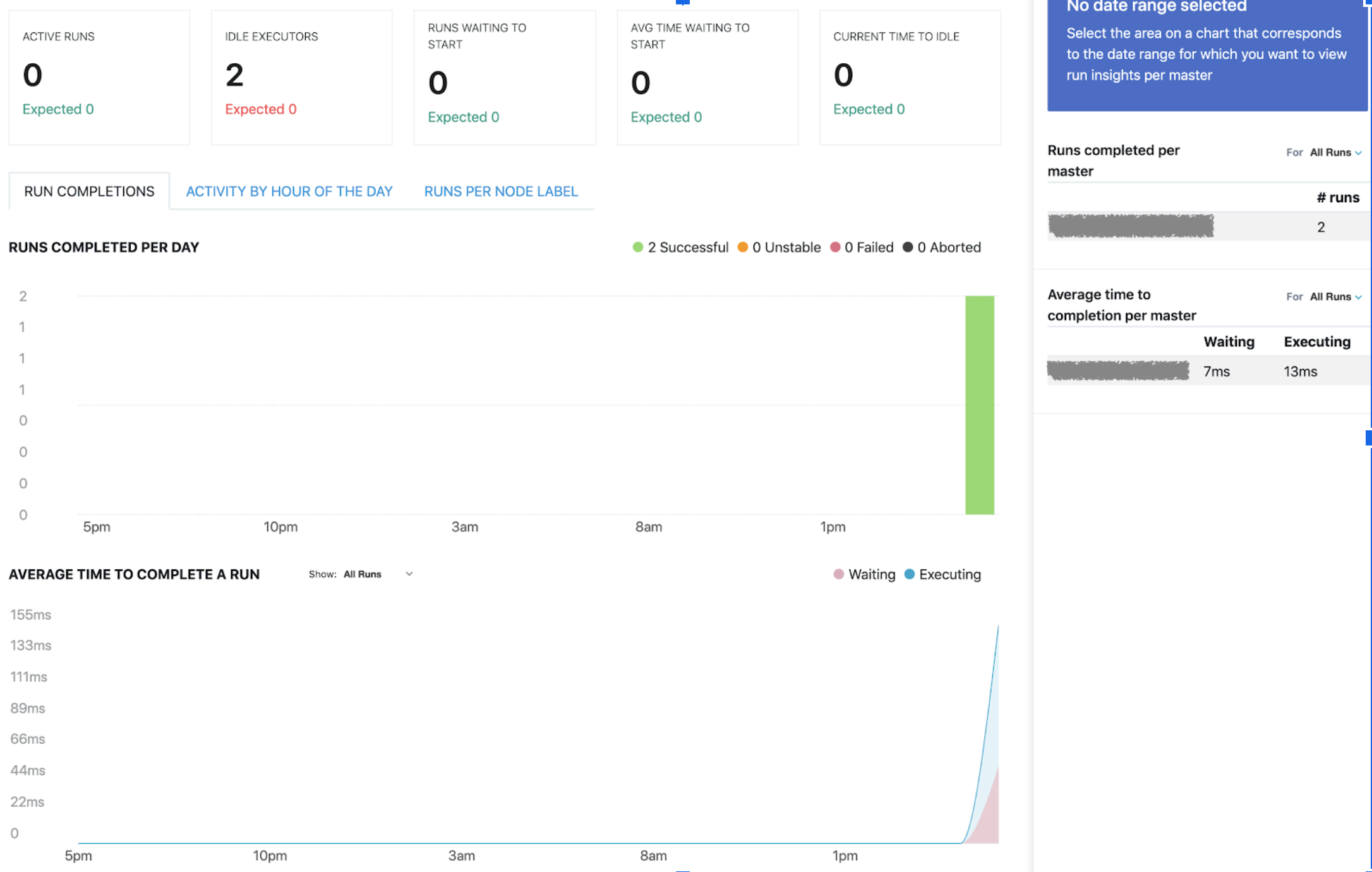
which is exactly what we expected.
So, there you go! Enjoy!
